After the outbreak of the novel coronavirus in 2020, vaccines, antibodies (Abs), small molecule drugs and other interventions need to be researched, and such a rapid development is partly due to the accumulation of scientific research in severe acute respiratory syndrome virus (SARS virus, SARS-CoV-1), MERS-CoV and other coronaviruses that cause human disease. SARS-CoV-1 and SARS-CoV-2 belong to the Sarbecovirus of the Beta coronavirus, and have similarities in the structure and function of the surface membrane protein Spike, and both invade cells mediated by the interaction between the viral spike (S) glycoprotein and the ACE2 receptor on the host cell surface. However, due to the limitations of technology and methodology, there exist many questions in the characteristics of SARS-CoV-1 antibody response and the relation to SARS-CoV-2 antibody response.
Professor ZHANG Linqi's team from School of Medicine, Tsinghua University, Professor LI Taisheng's team from Peking Union Medical College Hospital, and Professor YANG Yuhe's team from National Center for Nanoscience and Technology, published online in Immunity under the title "Dissecting the intricacies of human antibody responses to SARS-CoV-1 and SARS-CoV-2 infection". The antibody response of SARS-CoV-1 convalescents was analyzed from the aspects of polyclonal antibodies (pAbs) and monoclonal antibodies (mAbs), and was compared the duration, classification and properties of RBD Abs.
Using blood samples of SARS convalescents during their recovery period that had been stored for 20 years, the study systematically analyzed the antibody responses of SARS-CoV-1 patients, and made a comprehensive comparison with the antibody responses of SARS-CoV-2 patients. The plasma neutralization titer of SARS-CoV-1 convalescents was higher than that of SARS-CoV-2 severe convalescents, and the duration was longer. One reason may be the higher proportion of antibodies targeting SARS-CoV-1 RBD. In addition, the cross-binding of SARS-CoV-1 convalescents’ plasma at a high level, but the cross-neutralization is low, suggesting differences in spike gene sequence, protein structure and immunogenicity between the two viruses.
The team isolated 77 mAbs from SARS-CoV-1 convalescents, 60 of them target the RBD, mostly show strong neutralization with SARS-CoV-1, while the other 17 non-RBD Abs showed little or no neutralization. In addition, the neutralization proportion of these RBD Abs was significantly higher than that isolated from SARS-CoV-2 convalescents, consistent with the result of plasma antibodies response. The antibodies induced by two coronaviruses have similar high-frequency mutation after infection, indicating that the mutation and maturation rate of antibody genes follow the internal rhythm of the human body, and have no bearing on the type of virus or the infection severity. Then further analyzed the structure and function of 60 RBD antibodies. 60 RBD-targeting antibodies were classified into seven groups by competitive SPR analysis with ACE2 or typically antibodies with known epitopes, named RBD-1 to 7. RBD 1-3 are RBM antibodies, which compete strongly with ACE2 and 80R, have different competitive against S230, m396 and S309, respectively.
Then further characterized the binding modes and specificities of 21 selected Abs from the seven groups to the spike of SARS-CoV-1 by single-particle, negative-stain electron microscope (ns-EM). RBD-1 Abs bound to the "peak" of the RBD, RBD-2 Abs bound to the "mesa" and inner ridge of the RBM, RBD-3 Abs targeted to the outer ridge of RBM, RBD 1-3 Abs showed high neutralization to SARS-CoV-1 and weak cross activity. However, RDB-5 and RDB-6 Abs do not compete strongly with ACE2, and the binding sites mainly include the outer face (RDB-5) or the inner face (RDB-6) of RBD. RBD-6 Abs has the weak neutralization but the strong cross-reactivity, the IgG and Spike trimer complexes all form the dimers-of-trimers or trimers-of-trimers conformation. RBD-7 Abs target a specific region on the "up" RBD that interfaced with the NTD of the adjacent spike protomer (NTD proximal). Abs targeting this epitope is extremely rare in reported human COVID-19 convalescents, but it accounts for a large proportion (13/60) in SARS-CoV-1 convalescents, and has the same V region gene, which is one of the characteristics of antibody response in SARS-CoV-1 convalescents. Finally, the team evaluated the cross-neutralization of eight Abs against multiple novel coronavirus mutants and other ACE2 receptor coronaviruses, and analyzed the structure of the Spike protein complex of two cross-antibodies (W328-6A1 and W328-6E10) by cryo-electron microscopy(cryo-EM). It provides structural information for further understanding the broad neutralization mechanism.
“This work deep researches the SARS-CoV-1 antibodies field, and parallel research the antibody response to COVID-19, so we believe that this research perspective will be very valuable. The antibody response caused by SARS-CoV-1 and SARS-CoV-2 infection has many similarities, such as their antibodies can cross each other, and the RBD antibodies mainly contribute to the neutralization” ZHANG Linqi says, the senior author of the current work in Tsinghua University. “This study firstly classify RBD-targeting antibodies of SARS, provides important basis and guidance for the development of new-generation broad coronaviruses antibody drugs and vaccines” YANG Yuhe says, a Professor of NCNST and the senior author of the current work.
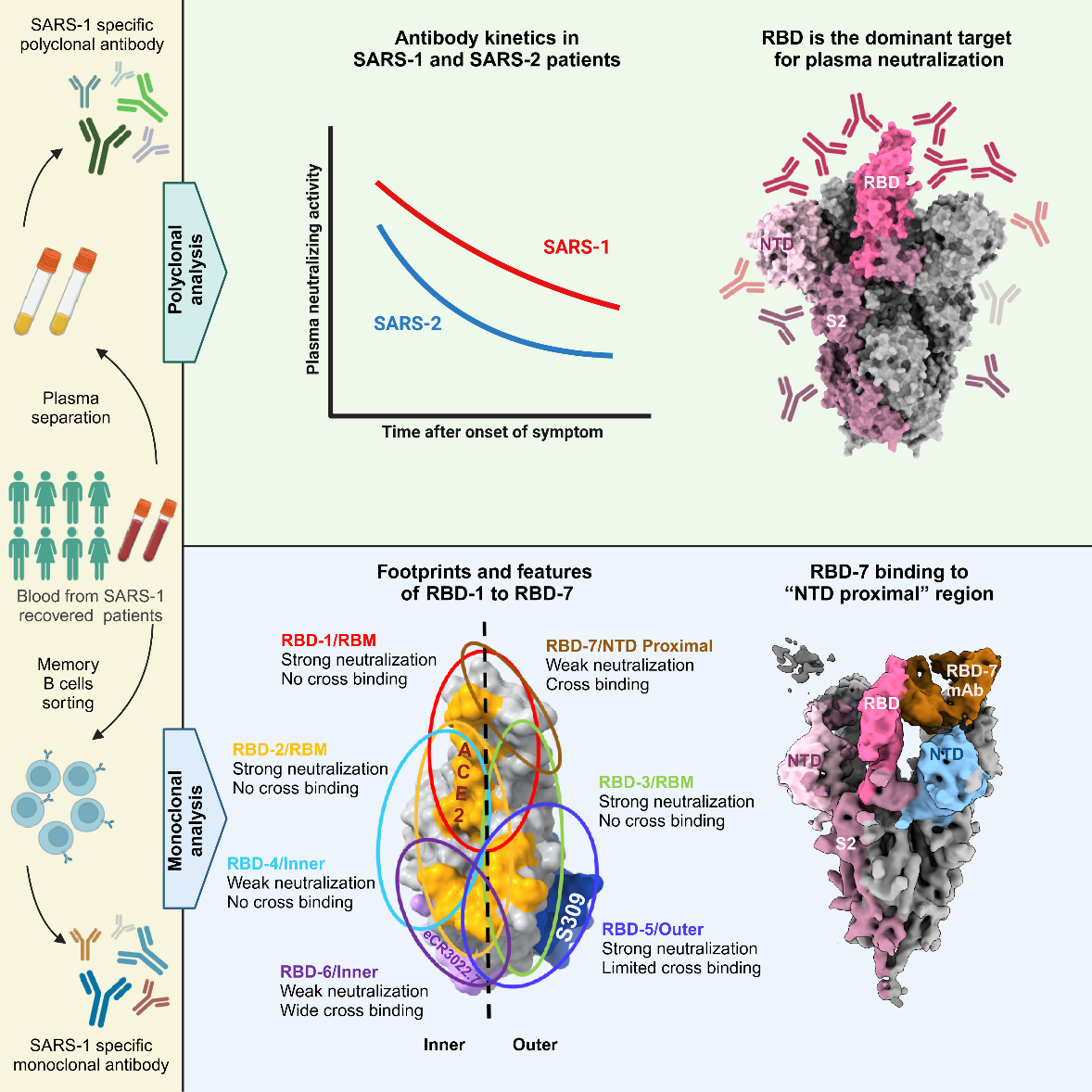
RBD mAbs form seven groups (RBD-1 to RBD-7) based on binding and structural profiles (Image by WANG et al)
Contact:
YANG Yuhe
National Center for Nanoscience and Technology (NCNST)
E-mail: yangyh@nanoctr.cn
LI Taisheng
Peking Union Medical College Hospital
E-mail: litsh@263.net
ZHANG Linqi
School of Medicine, Tsinghua University
E-mail: zhanglinqi@tsinghua.edu.cn




