Recently, the research team led by SHI Xinghua from the National Center for Nanoscience and Technology (NCNST) of the Chinese Academy of Sciences (CAS), in collaboration with GAO Huajian's team from Tsinghua University, developed a new reinforcement learning-based enhanced sampling method called Adaptive Collective Variables Generator (Adaptive CVgen). This method has been successfully applied to study protein folding and the synthesis of fullerene (C60). The study was published in the Proceedings of the National Academy of Sciences (PNAS).
One of the biggest challenges in fundamental research is understanding the dynamic evolution of microscopic systems, which plays a key role in areas like protein folding, drug development, and materials design. Current experimental techniques are limited in capturing these dynamic processes. For example, cryo-electron microscopy can resolve static protein structures but cannot reveal the transient, dynamic changes. In contrast, computational methods have shown potential in exploring these behaviors. Current tools mainly include deep learning-based structure prediction, molecular dynamics simulations, and enhanced sampling methods for long-timescale simulations.
Deep learning tools like AlphaFold, which won this year's Nobel Prize in Chemistry, can predict protein structures but not their folding dynamics. Traditional molecular dynamics methods are limited to short timescales, making them effective only near equilibrium states. Enhanced sampling methods, which can handle long timescales, are promising but have mainly been applied to simple systems. There’s an urgent need for enhanced sampling methods with broader applicability for more complex challenges.
Adaptive CVgen is an advanced adaptive sampling technique developed to preserve the free energy landscape of molecular systems in simulations, allowing for accurate capturing of both thermodynamic and dynamic details. The method is characterized by two core innovations: high-dimensional collective variables (CVs) and reinforcement learning-driven predictive capabilities. By constructing an extensive set of CVs that spans all possible conformational states, Adaptive CVgen offers a precise and localized view of system evolution. Unlike traditional global approaches, which may miss specific structural variations, this method’s high-dimensional CVs detect minute local changes, representing detailed features with clarity. Additionally, Adaptive CVgen incorporates long-range correlations within the CV framework to account for interactions at varied distances and across different regions in the system, thus supporting a comprehensive view of complex dynamics. Reinforcement learning further enhances the method by iteratively refining CVs based on historical trajectory data, allowing for adaptive predictions of the system’s evolutionary path and extensive exploration of conformational space.
The Adaptive CVgen workflow follows a structured, iterative process comprising four main steps: generating high-dimensional CVs, running simulations, updating CV weights, and selecting optimal conformations for next round. Initially, a diverse set of CVs is constructed to cover the entire spectrum of the system's behaviors. In each round of simulation, new simulations are launched from selected conformations, with multiple parallel replicas running to ensure robust data collection. Following each round, reinforcement learning updates the CV weights based on accumulated trajectory data, iteratively refining the exploration of potential system evolutions. This process repeats—simulation, CV weight adjustments, and optimal conformation selection—until the system achieves convergence, enabling a thorough and adaptive mapping of the conformational landscape.
The Adaptive CVgen has shown considerable success in diverse applications, particularly in the study of protein folding and chemical synthesis. In protein folding studies, Adaptive CVgen has been applied across various proteins with multiple secondary structures, capturing dynamic folding processes without requiring parameter adjustments. It has also been employed to model chemical reactions, such as fullerene synthesis, successfully simulating intermediate steps and achieving final structures identical to standard fullerenes. Such applications demonstrate the method’s versatility and its ability to address complex systems that challenge traditional low-dimensional approaches. Notably, the systems studied in this research pose significant challenges for existing techniques, representing a major advancement in long-timescale simulation research.
“Adaptive CVgen also has broad potential for applications in biocatalysis, gene expression and regulation, drug development, chemical synthesis, catalytic reactions, and materials engineering. Its further development could significantly advance research in the dynamics of complex systems, offering new insights and tools.” SHI Xinghua says, a Professor of NCNST and the senior author of the current work.
Contact: SHI Xinghua
National Center for Nanoscience and Technology (NCNST)
E-mail: shixh@nanoctr.cn
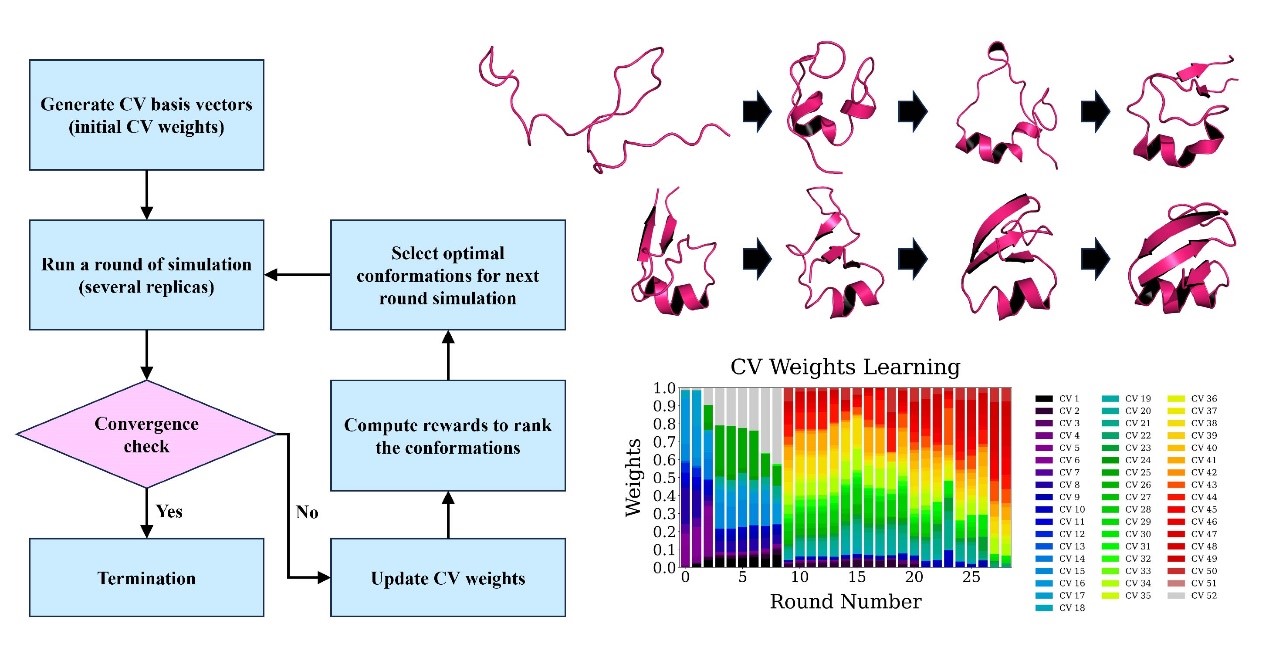
Figure 1. Methodology flowchart of Adaptive CVgen. (Image by SHI Xinghua et al)
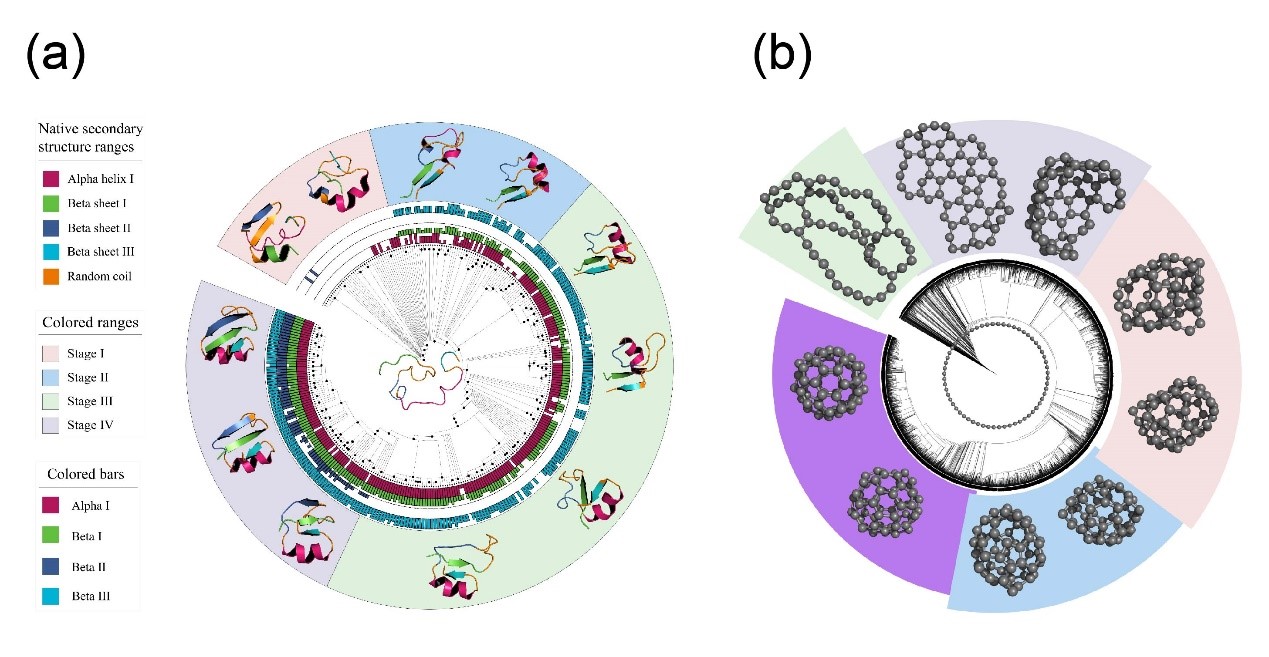
Figure 2.Structure evolution sampled by Adaptive CVgen. (a) Protein folding process (PDB ID: 2hba), illustrating the transition from disorder to order and instability to stability, revealing key dynamics in protein folding. (b) Fullerene (C60) synthesis pathway, demonstrating the transformation from 2D to 3D structures and highlighting the crucial steps in C60 synthesis. (Image by SHI Xinghua et al)




